
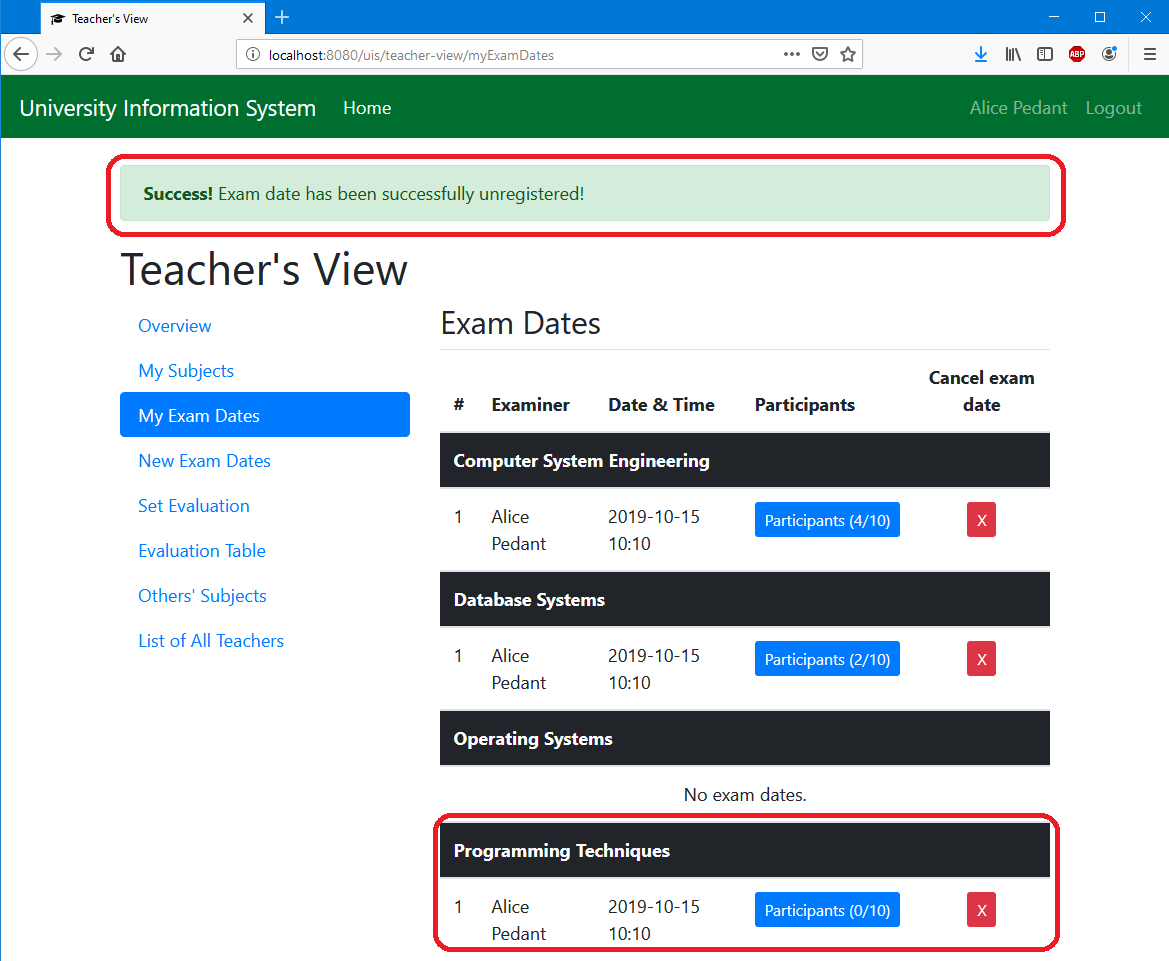
Has been responsible for our understanding of many biological processes and is an excellent method for identifying genes that Of the mutant phenotype coupled with molecular analyses of the gene allows elucidation of the gene's function. After a mutant is found, the gene mutated is identified through standard molecular techniques. In classicalįorward genetic screening, individuals are treated with mutagens to induce DNA lesions and mutants with a phenotype of interestĪre sought. Through genetic analyses, the function of genes is investigated by studying organisms where gene function is altered. Referencesġ. Introduction to reverse genetics in C. Recovering, genotyping, and backcrossing a live mutant animal 7.5. Protocols for constructing the frozen library of mutagenized animals and DNA templates 7.3.

Construction and screening of deletion mutant libraries to generate C. Feeding two bacterial strains at the same time 6.5. RNAi feeding in liquid culture (96 well format) 6.1. Feeding two bacterial strains at the same time 5.5. Preparing template for in vitro transcription 3.2. Did I target the gene I wanted to target? 3. Desigining a large-scale RNAi screen 2.7. Can I use RNAi to target multiple genes? 2.6. How do I know whether the RNAi has worked? 2.5. Using RNAi to knockdown gene function 2.1. ( 14) does not contain sufficient information to enable us to make quantitative comparisons of our methods nor have we found their procedure used in the published literature.Table of Contents 1. We have experienced difficulty with protocols incorporating such methods and have avoided them in RICH. Moreover, EL-CSC uses physical trapping through biotin–avidin complex formation to enrich for products. In our procedure, we use a selection adaptor that allows us to generate an RNA intermediate, and so we can isolate heteroduplexes that have formed at only one end. Thus, cloning of cDNA fragments that span introns is much less likely. Unlike our procedure, their method requires heteroduplex formation at both ends between cDNA and genomic fragments, because both ends of the heteroduplex must be ligated. Like ours, their protocol is based upon heteroduplex formation between DNA fragments made from two populations, and the use of ligation (with what they call “capture oligonucleotides”) to distinguish heteroduplex from homoduplex. This method should work whenever a cDNA population is available that contains transcripts from the gene in question.Ī protocol (end ligation coincident sequence cloning, EL-CSC) similar to ours, has been presented by Brookes et al. We have described an additional approach to this problem: an effective protocol for selecting cDNA fragments that are homologous at one of their ends to one of the ends from a collection of genomic fragments. Direct hybridization selection ( 1) has also found use, but it diminishes in usefulness with rare messages and suffers from the vagaries of physical selection methods and background problems with repetitive sequences.
But DNA sequencing on a massive scale is still costly. Even without homology, computational methods for predicting genes also have promise ( 4). DNA sequencing is effective when the gene in question has homology to a known expressed sequence. But it fails when introns are absent or the intron–exon borders are not recognized and exon trapping yields a background of false candidates derived from cryptic splice sites. Exon trapping works when the gene in question contains splicing sites that are efficiently recognized by the host cell ( 6– 8). RNA transcription and attenuation.įinding genes in large chromosomal regions has been approached in three ways: exon trapping, DNA sequencing analysis, and direct hybridization selection. The DNA is dissolved in 25 μl of RNase-free water after washing with 70% ethanol. The PCR products are purified with phenol/chloroform extraction and ethanol precipitation. After initial denaturation of 94☌ for 5 min, 2.5 units of Pfu polymerase is added to the mixture and 20 cycles at 94☌ for 1 min and 70☌ for 4 min are performed, followed by a final extension at 72☌ for 10 min. The 100-μl PCR mixture contains 8 μl of the DNA, all four dNTPs (each at 200 μM), 1 μM pN-24, 1 μM SP6, and Pfu buffer at a 1× final concentration. Eighty microliters of TE is added to the reaction mixture and incubated in boiling water for 5 min. After purification with S-300 HR columns, the DNA is digested at 37☌ with 10 units of λ exonuclease. The DNA is amplified by a PCR program that specifies an incubation for 10 min at 94☌ followed by 10 cycles of 1 min at 94☌, 1 min at 60☌, and 3 min at 72☌. Five microliters of the DNA solution (component E) is mixed with 50 μl of reaction mixture.


 0 kommentar(er)
0 kommentar(er)
